Autophagyis crucial for maintaining cellular homeostasis. It can be divided intonon-selective autophagy and selective autophagy, both of which exertsignificant functions. Mitophagy, ribosomal autophagy, endoplasmic reticulumautophagy, and peroxisomal autophagy all belong to selective autophagy.Selective autophagy is closely associated with major human diseases, such asneurodegenerative diseases and metabolic diseases. Clarifying molecularmechanisms of selective autophagy can provide a theoretical basis for thedevelopment of related drugs or other treatments.
Inselective autophagy, cargo receptors recognize specific targets withdegradation signals and interact with LC3 (Atg8 in yeast and plant cells).Atg11 with four coil-coiled domains (i.e. CC1, CC2, CC3 and CC4) exhibits as ascaffold protein in almost all types of selective autophagy, which isconsidered a marker protein for selective autophagy. It binds to substratereceptors through the CC4 domain, when interacting with the autophagy coreprotein Atg1 through the CC2 and CC3 domains, thereby recruiting substrates toautophagosomes. Though part of Atg11’s roles in selective autophagy have beenrevealed, how Atg11 activity is regulated remains to be explored by furtherinvestigation.
OnApril 15, the research group led by Prof. YI Cong from the Zhejiang UniversitySchool of Basic Medicine/the First Affiliated Hospital of the ZhejiangUniversity School of Medicine, published an article entitled Atg1-mediatedAtg11 phosphorylation is required for selective autophagy by regulatingautophagy in the journal Autophagy, uncovering a new mechanism by which Atg11regulates selective autophagy.
Inthis study, the researchers found that Atg11 was phosphorylated when yeastcells were subjected to rapamycin treatment, nitrogen starvation, or glucosestarvation. Next, the group discovered that Atg11 is a direct substrate of thekinase Atg1. In vitro phosphorylation experiments revealed that the Atg11 CC4domain, which is responsible for binding to the receptor protein, wasphosphorylated by Atg1. The results strongly suggested that the process ofAtg11 binding to selective autophagy receptors is regulated by Atg1 kinaseactivity. In order to identify its phosphorylation sites, the Atg11 CC4obtained from in vitro phosphorylation experiments was analyzed by massspectrometry, and S949, S1057, and S1064 were identified as the phosphorylationsites of Atg1-mediated phosphorylation of Atg11. Further functional analysisexperiments showed that mutations at these three sites curb selective autophagydue to the incapability of Atg11 binding to selective autophagy receptors(Figure 1).
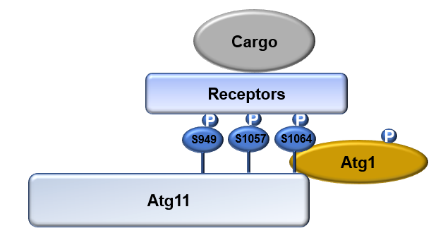
Figure1. Model for the regulation of selective autophagy by Atg1-mediated Atg11phosphorylation
Inconclusion, Atg11 regulates the progression of selective autophagy viaswitching its phosphorylation status then affecting its binding ability toselective autophagy receptors. This discovery expands our understanding of theregulatory mechanism underlying the binding between Atg11 and selectivereceptors. As one of the reviewers said, I find this study reveals a new layerof regulation in selective autophagy.












