Oxidative phosphorylation is the major cellular process utilized by most free-living eukaryotes to gain energy, in the form of ATP, from catabolism. It happens on the inner mitochondria membrane (IMM), where four membrane-bound complexes (complex I-IV, CI-CIV) first transport electron from the reductive metabolite NADH to molecular oxygen O2. Current structural studies of the above electron transport chain (ETC) complexes are limited to yeast, mammals, algae and vascular plant, belonging to the Opisthokonta and Archaeplastida clades of eukaryotes. For other eukaryotic clades such as the Alveolates, although tomographic studies have already revealed diversities in their mitochondrial cristae morphologies, structural investigations into the ETC complexes are essentially lacking.
On March 31st 2022, Dr. ZHOU Long from the Zhejiang University School of Medicine and Dr.James Letts from the Molecular and Cellular Biology Department at University of California Davis collaborated to publish a research article in the journal Science. The study reported cryoEM structures of a 2.3 MDa super-complex I+III2 (SC I+III2) and a 2.7 MDa complex IV dimer (CIV2) of Tetrahymena thermophila (Tt), a key ciliate model organism belonging to the Alveolata clade, at 2.6 Å and 3.0 Å resolution respectively. Compared to the 1.4 MDa mammalian SC I+III2 with 67 subunits, Tt-SC I+III2 encompasses 91 subunits, out of which 20 subunits have no reported homologues. Moreover, the Tt-CIV2 has 52 subunits per protomer and has surpassed CI as the largest individual ETC complexes, while existing eukaryotic CIV structures usually consist of 10-14 subunits with ~200 KDa mass.
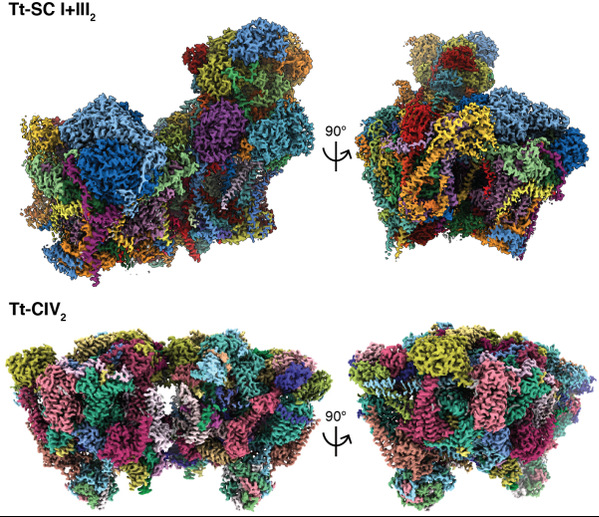
Instead of the ‘14 core subunit’
architecture shared by most species’ CI, Tt-CI has three core subunits ND1, ND2
and ND5 split into two poly-peptide chains. Neither the usual ‘active-deactive’
activity transition nor the ‘open-close’ conformational change exists in Tt-CI,
while local conformations of the loop regions near ubiquinone/co-enzyme Q (CoQ)
tunnel resemble mammalian CI in the closed state. These detailed structural
information of the constitutively active CI suggests that ‘open-close’ state
transition as well as local CoQ loop rearrangements may not be required by the
conserved mechanism of CI substrate turnover and redox-H+ pumping coupling.
Tt-CI actually merges with Tt-CIII2 into an
obligatory SC I+III2, the tight association pulls the two complexes closer in
the inter-membrane space (IMS), bending the joint membrane domain to fit the
curvature of Tetrahymena’s cristae IMM. This likely leads to the high degree of
symmetry-breaking between the two CIII protomers, as the inter-heme bL distance
has increased from the highly conserved 11 Å to 14 Å, beyond the direct
electron transfer distance. The physiological role of SC I+III2 formation is
thus indicated to be the specialization of the two CIII2 CoQ cavities and
turnover rate coupling between CI and CIII2.
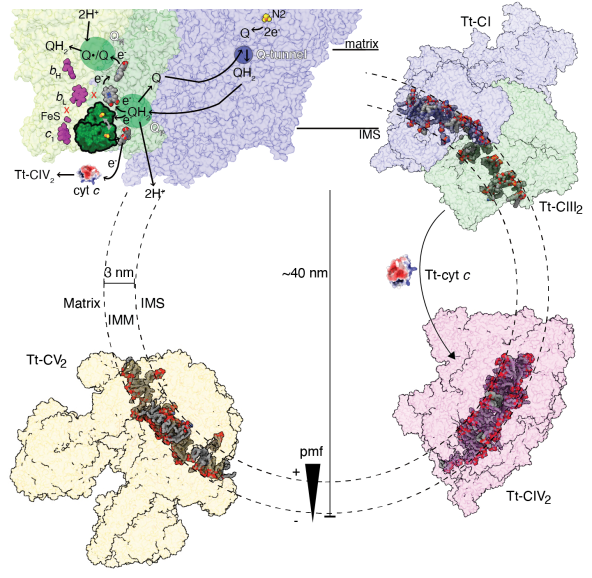
Tt-CIV2 is highly divergent as compared to
the classic 14 subunit mammalian model. Its core subunit extensions and unique
accessory subunits form a cytochrome c binding crate with positive surface
electrostatic potential, while homology modeling shows that CIV binding site of
Tt cytochrome c is negatively charged. Such swapped charge as compared to
mammalian CIV-cytochrome c interactions explains why Tt-CIV can’t oxidase
mammalian cytochrome c. In addition, Tt-CIV2 also harbors three mitochondrial
carriers belonging to the solute carrier family 25A (SLC25A), as well as a
hexameric α-propeller domain similar to the transporting chaperone formed by
translocase of the inner membrane (TIM) 8, 9, 10 or 12. All these non-electron
transporting accessory subunits suggest that Tt-CIV2 is more than a ETC
complex, but has evolved into a mitochondrial redox homeostasis and
cross-membrane translocation hub in ciliates.
“People usually think of
eukaryotes as a taxonomic combination of yeast, C. elegans, drosophila,
zebrafish and mice, but there are much more populations than those. This work
has certainly demonstrated a higher level of diversity in eukaryotic ETC
structures than we used to believe, thus opened up many new possibilities in
the field of bioenergetics. Investigation into protist ETC structures and the
approach of solving multiple complex structures from one cryoEM dataset also
provided new angles into the pharmaceutical developments against
life-threatening parasites including plasmodium and leishmania.”Dr. Zhou said.










